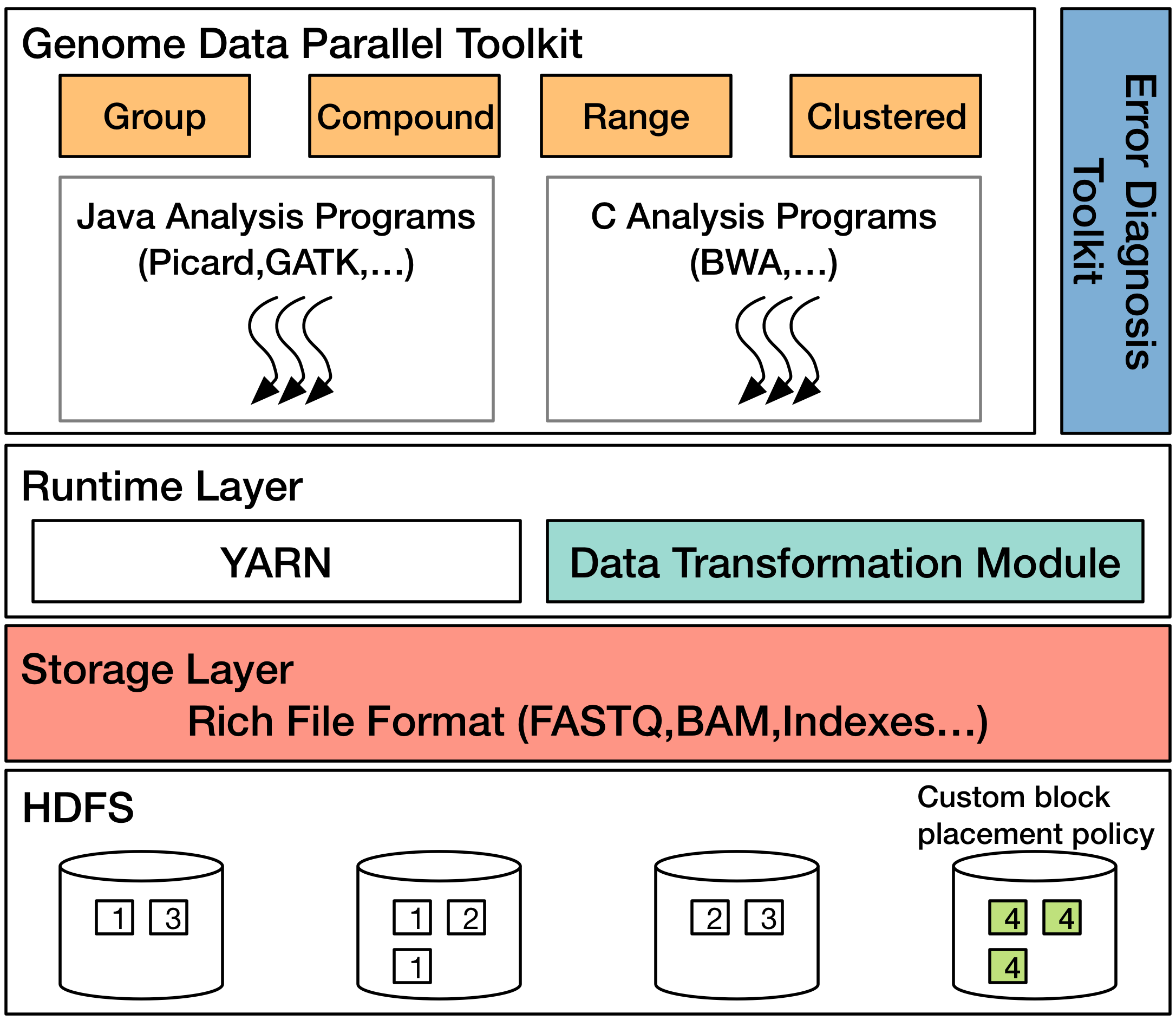
Overview
Today large sequencing centers are producing genomic data at the rate of 10 terabytes a day and require complicated processing to transform massive amounts of noisy data into biological information. To address these needs, we are developing GESALL (GEnomic Scalable Analysis with Low Latency), a system for end-to-end processing of the genomic data. We aim to improve the overall system performance by using a variety of ideas from the database systems research community.
Publications
-
Parallel Processing of Windowed Variant Callers. UMass CICS Technical Report (UM-CS-2018-006).
-
Abhishek Roy, Yanlei Diao, Uday Evani, Avinash Abhyankar, Clinton Howarth, Rémi Le Priol, Toby Bloom. Massively Parallel Processing of Whole Genome Sequence Data: An In-Depth Performance Study. In ACM SIGMOD International Conference on Management of Data (SIGMOD 2017). [pdf]
-
Yanlei Diao, Abhishek Roy, Toby Bloom. Building Highly-Optimized, Low-Latency Pipelines for Genomic Data Analysis. In 7th Biennial Conference on Innovative Data Systems Research (CIDR 2015). [pdf]
-
Abhishek Roy, Yanlei Diao, Evan Mauceli, Yiping Shen, Bai-Lin Wu. Massive Genomic Data Processing and Deep Analysis. In Proceedings of the VLDB Endowment 5(12) (VLDB 2012). [pdf]
Downloads
GESALL code repository is hosted at GitLab - https://gitlab.com/gesall
People
GESALL is being developed by Yanlei Diao, Abhishek Roy, and Prashant Shenoy at the University of Massachusetts Amherst. Our collaborators include Evan Mauceli, Dr. Yiping Shen, Dr. Bai-Lin Wu at the Children's Hospital Boston and Uday Evani, Avinash Abhyankar, Dr. Toby Bloom at the New York Genome Center.
Support
We gratefully acknowledge the funding provided by the following agencies:

National Science Foundation
DBI-1356486
Massachusetts Green High Performance Computing Center
